Abstract
Tumor-necrosis factor (TNF), a pleiotropic cytokine, triggers physiological and pathological responses in several organs. Here we show that deletion of the mouse gene Timp3 resulted in an increase in TNF-α converting enzyme activity, constitutive release of TNF and activation of TNF signaling in the liver. The increase in TNF in Timp3−/− mice culminated in hepatic lymphocyte infiltration and necrosis, features that are also seen in chronic active hepatitis in humans. This pathology was prevented when deletion of Timp3 was combined with Tnfrsf1a deficiency. In a liver regeneration model that requires TNF signaling, Timp3−/− mice succumbed to liver failure. Hepatocytes from Timp3−/− mice completed the cell cycle but then underwent cell death owing to sustained activation of TNF. This hepatocyte cell death was completely rescued by a neutralizing antibody to TNF. Dysregulation of TNF occurred specifically in Timp3−/−, and not Timp1−/− mice. These data indicate that TIMP3 is a crucial innate negative regulator of TNF in both tissue homeostasis and tissue response to injury.
Similar content being viewed by others
Main
Mutation of TIMP3, which encodes a member of the tissue inhibitor of metalloproteinase (TIMP) family, results in the human retinal degenerative disease Sorsby's fundus dystrophy (SFD)1. Deletion of Timp3 in mice results in spontaneous pulmonary air space enlargement and enhanced mammary epithelial apoptosis2,3. Biochemical studies suggest that TIMP3 is unique in its ability to inhibit TNF-α converting enzyme (TACE; also known as disintegrin and metalloproteinase domain 17 or ADAM17; refs. 4,5). Deletion of the gene encoding TACE, Adam17, results in the defective release of membrane-bound TNF6,7, and TACE is important for shedding several ligands and receptors8,9,10,11,12. TNF has a central role in physiological processes such as immunity and organ regeneration, as well as in diseases including rheumatoid arthritis, diabetes, chronic inflammation and autoimmune disorders13,14.
We investigated whether deletion of Timp3 alters TNF bioactivity and signaling in vivo. Our results show that TIMP3 controls TNF and, as a consequence, affects inflammation and organ regeneration.
Results
Constitutive TNF shedding in Timp3−/− mice
We first examined several organs to determine whether basal TNF levels were altered in Timp3−/− mice. TNF is normally undetectable in the liver, spleen, lung, brain, serum (Fig. 1a) and kidney (data not shown) of wild-type mice. In Timp3−/− mice, TNF was measurable only in the liver (Fig. 1a). We detected an increase in TNF by enzyme-linked immunosorbent assay (ELISA) in liver protein homogenates of Timp3−/− mice, which was confirmed by an increase in hepatic TNF detected immunohistochemically. Immunohistochemical analysis also localized TNF to the hepatic sinusoids (Fig. 1b,c). In the sinusoids, TNF immunostaining colocalized with Kupffer cells, the resident macrophages of the liver, as shown by immunodetection with antibodies to F4/80 in adjacent sections (Fig. 1d,e).
(a) Quantification of TNF by ELISA in organs of 12-week-old wild-type (+/+) and Timp3−/− (−/−) mice. *P < 0.05. (a,b) Immunohistochemical staining for TNF in livers of wild-type (b) and Timp3−/− (c) mice. (e,f) Adjacent sections immunostained for the macrophage-specific marker F4/80 in livers of wild-type (d) and Timp3−/− (e) mice. (f) Appearance of soluble TNF (17 kDa) in livers of Timp3−/− mice (100 μg of protein loaded per lane). Soluble recombinant TNF (rTNF) was used as a positive control. Membrane-bound TNF is visualized as a 26-kDa band. (g–j) In situ hybridization on livers of wild-type mice with Timp3 sense and antisense riboprobes. (k) Quantitative real-time TaqMan RT-PCR of Timp3 mRNA in livers of wild-type mice (n = 3) and Timp3−/− mice with (n = 3) and without (n = 2) bone marrow transplantation (BMT) of GFP-tagged wild-type bone marrow. Data are the mean ± s.e.m. Scale bars: b–e, 40 μm; insets in d,e, 0.1 mm; g,h, 50 μm.
Histomorphometric quantification showed that the number of F4/80-positive cells was not altered in Timp3−/− mice (data not shown), suggesting that the increase in TNF might be due to an increase in the production or release of this cytokine by Kupffer cells. Membrane-bound TNF is a 26-kDa protein that is proteolytically processed into a 17-kDa soluble form15,16. Western blotting showed that there was more of both forms of TNF, but particularly soluble TNF, in the livers of Timp3−/− mice compared with those of wild-type mice (Fig. 1f). By real-time TaqMan RT-PCR, we found no difference in the expression of Tnf mRNA in livers of wild-type and Timp3−/− mice (data not shown).
We sought to confirm the presence and localization of Timp3 expression in livers of wild-type mice. Timp3 mRNA is abundant in mouse liver17 and is primarily expressed in hepatocytes18. In situ hybridization on wild-type liver tissue showed that Timp3 mRNA localized to hepatocytes (Fig. 1g–j). Because hepatocytes constitute ∼95% of the liver volume, it is difficult to attribute Timp3 expression to smaller liver cell populations, such as Kupffer cells. We used bone marrow transplantation to test for the production of Timp3 mRNA by the intrahepatic bone marrow–derived compartment that includes Kupffer cells19. Timp3−/− mice that received transplants of green fluorescent protein (GFP)-labeled wild-type bone marrow showed detectable expression of Timp3 in the liver by real-time TaqMan RT-PCR, indicating that intrahepatic bone marrow–derived cells could express Timp3 (Fig. 1k). As expected, this level of Timp3 expression was 100 times lower than that found in livers of wild-type mice, where hepatocytes also contributed to the production of Timp3 (Fig. 1k).
Increased TACE activity in livers of Timp3−/− mice
Next, we used a fluorogenic TACE peptide substrate to determine TACE activity and found that cleavage was 1.8 times higher in liver extracts from Timp3−/− mice than in those from their control littermates (Fig. 2a). Adding recombinant TIMP3, but not recombinant TIMP1, to liver extracts reduced this enzymatic activity (Fig. 2a). This differential response to TIMP3 is the signature of TACE activity4. Adding the serine protease inhibitor aprotinin or the matrix metalloproteinase (MMP)-specific inhibitor BMS-275291 (ref. 20) did not reduce the increase in enzymatic activity in liver extracts from Timp3−/− mice (Fig. 2a).
(a) Fluorometric measurements of TACE peptide cleavage in livers of 12-week-old wild-type and Timp3−/− mice in the absence of any inhibitors or in the presence of recombinant TIMP1 (rTIMP1; 100 nM), recombinant TIMP3 (rTIMP3; 100 nM), aprotinin (5 μg ml−1) or BMS-275291 (1 μM). Data are the mean ± s.e.m. (n = 3 mice for each genotype). *P < 0.05. (b) Cleavage of TACE peptide substrate by 50 nM recombinant TACE (rTACE), recombinant MMP14 (rMMP14) or recombinant MMP17 (rMMP17), in the absence or presence of 100 nM recombinant TIMP1 (rTIMP1) or TIMP3 (rTIMP3). (c–e) Fluorometric measurements indicating cleavage of GLARE (c), gelatin (d) or fibronectin (e) in livers of wild-type and Timp3−/− mice. Data are the mean ± s.e.m. (n = 3 mice per genotype). As a positive control, 100 nM recombinant MMP7 (rMMP7), MMP14 (rMMP14) or MMP17 (rMMP17) was added in these assays.
The membrane-type MMPs MMP14 and MMP17 have TNF convertase activity in vitro21,22. We found that only recombinant TACE, and not MMP14 or MMP17, efficiently cleaved the fluorogenic TACE peptide substrate, verifying the specificity of this cleavage assay for TACE (Fig. 2b). As above, recombinant TACE activity was inhibited only by recombinant TIMP3 and not by recombinant TIMP1 (Fig. 2b). MMP7 can also shed TNF in vivo23. Using an MMP7 peptide substrate (GLARE), we ruled out any increase in MMP7 activity in the liver as a result of Timp3 deficiency (Fig. 2c). In fact, MMP7 was undetectable by immunoblotting in livers of both wild-type and Timp3−/− mice (data not shown). In addition, we detected no increase in general MMP activity in two independent assays that used either gelatin or fibronectin as substrates (Fig. 2d,e).
To confirm the ability of these MMP activity assays to detect MMP14 and MMP17, we added the recombinant proteins to GLARE, gelatin and fibronectin substrates. Both GLARE and gelatin were cleaved by MMP14 and, to a lesser extent, by MMP17 (Fig. 2c–e), verifying the sensitivity of the assays to these membrane-type MMPs. Real-time TaqMan RT-PCR analyses also showed that there was no difference in the expression of Adam17 mRNA in livers of wild-type and Timp3−/− mice. Taken together, these results indicate that livers of Timp3−/− mice have increased TACE activity but no difference in MMP activity.
Activation of TNF signaling in livers of Timp3−/− mice
Activation of the transcription factor NF-κB, a heterodimer composed of p65 (also called RelA), RelB or cRel and p50 or p52 subunits24, is central to TNF signaling. In the liver, NF-κB is normally present as the p65-p50 heterodimer. Notably, loss of p65 in mice blocks NF-κB activation, resulting in embryonic lethality owing to massive TNF-induced apoptosis in the liver25. These mice can be rescued by deleting the gene Tnfrsf1a, which encodes TNF receptor 1 (ref. 26). When NF-κB is activated, p65 is translocated from the cytoplasm to the nucleus.
To investigate whether an excess of TNF resulted in increased activation of NF-κB in livers of Timp3−/− mice, we examined the localization of p65 by immunohistochemistry. In the absence of TIMP3, p65 showed a preferentially nuclear localization, in contrast to its cytoplasmic granular appearance in age-matched wild-type controls (Fig. 3a–d). We quantified the cells showing nuclear localization of p65 by counting five random fields in pericentral regions of the liver at × 40 magnification. The number of cells with nuclear NF-κB localization was more than twice as high in livers of Timp3−/− mice (45.2 ± 6.6%; mean ± s.e.m.) versus wild-type mice (18.6 ± 4.9%; Fig. 3e).
(a–d) Representative images at low and high magnification of immunohistochemical staining for p65 in livers of wild-type (a,b) and Timp3−/− (c,d) mice. Arrowheads in d indicate positive nuclei. (e) Histomorphometric quantification (original magnification, × 40) of positive nuclei in pericentral hepatocytes of wild-type and Timp3−/− mice. (f) Quantification of IL-6 by ELISA in livers of wild-type and Timp3−/− mice. (g–j) Immunohistochemical staining for IL-6 in liver sections from wild-type (g,h) and Timp3−/− (i,j) mice. Arrowheads indicate positive staining. Data are the mean ± s.e.m. (n = 3 mice per group). *P < 0.05. Scale bars: a,c,h,j, 0.1 mm; b,d,g,i, 40 μm; h,j,insets, 50 μm.
We confirmed activation of the TNF signaling pathway by assessing the expression of interleukin-6 (IL-6), which is a transcriptional target of NF-κB27. Hepatic quantities of IL-6 measured by ELISA were 2.6 times higher in livers of Timp3−/− mice (Fig. 3f). Expression of IL-6 in Timp3−/− mice was localized to the Kupffer cells and to the cytoplasm of pericentral hepatocytes (Fig. 3g–j). Thus, in the absence of TIMP3, a low concentration of TNF activated the TNF signaling pathway in the liver. Notably, immunostaining of both p65 and IL-6 was most prominent in the pericentral regions.
Hepatic inflammation with aging in Timp3−/− mice
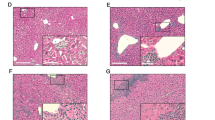
Given the aberrant activation of TNF signaling in livers of Timp3−/− mice, we determined whether the mutant mice had associated liver pathology. At 18–22 months of age, periportal lymphocytic infiltrates progressively appeared in the livers of Timp3−/− mice, with substantial alteration of the periportal lobular architecture (Fig. 4a,b). This was not observed in age-matched Timp3+/− mice (Fig. 4c). Hepatic necrosis was also evident in some areas (Fig. 4b). We also looked for evidence of hepatic fibrosis by trichrome staining in the livers of Timp3−/− mice but found no excessive deposition of collagen (Fig. 4d).
(a–c) Hematoxylin and eosin staining of liver sections from Timp3−/− (a,b) and Timp3+/− (c) mice. Arrowheads mark the borders of lymphocyte-infiltrated areas and the asterisk indicates hepatocyte necrosis. (d) Trichrome staining of liver from Timp3−/− mice. (e–j) Immunohistochemical detection of CD3 (e), B220 (f), CD4 (g), CD8 (h), neutrophils (i) and IFN-γ (j) in livers of Timp3−/− mice. Arrowheads indicate areas of positive stain. (k,l) Hematoxylin and eosin staining of liver sections from Tnfrsf1a−/− Timp3−/− (k) and Tnfrsf1a+/− Timp3−/− (l) mice. Arrowheads mark the borders of lymphocyte-infiltrated areas. Images are representative of six 22-month-old mice of each genotype. Scale bars, 0.1 mm.
Characterization of the lymphocytic infiltration showed that it comprised cytologically mature lymphocytes of a non-neoplastic nature. These hepatic infiltrates predominantly contained CD3+ T cells with occasional foci of B220+ B cells (Fig. 4e,f). Further characterization indicated that the T-cell population comprised mostly CD4+ cells and few CD8+ cells (Fig. 4g,h). A mouse neutrophil-specific antibody showed that neutrophils were occasionally present in the infiltrates (Fig. 4i). There was no evidence of cell turnover in these infiltrates, as assessed by measuring apoptosis by TUNEL and cell proliferation by immunohistochemistry of phosphorylated histone H3 (data not shown). The lack of cell turnover and presence of immunohistochemical heterogeneity confirmed that these mononuclear infiltrates are not clonogenic or tumoral.
Lymphocyte infiltration and hepatic necrosis are associated with chronic active hepatitis in humans28. They have been also reported in transgenic mice overexpressing interferon-γ (IFN-γ)29. We therefore determined whether IFN-γ was induced in the livers of aged Timp3−/− mice. We detected lymphoid cells immunopositive for IFN-γ in the periphery of these infiltrates (Fig. 4j). To confirm that TNF signaling was responsible for hepatic inflammation in Timp3−/− mice, we generated mice deficient in both Timp3 and Tnfrsf1a. We found no evidence of lymphocytic infiltration or hepatic necrosis in the livers of Tnfrsf1a−/− Timp3−/−mice at 18–22 months of age (Fig. 4k) compared with age-matched Tnfrsf1a+/− Timp3−/− mice (Fig. 4l). Thus, blocking TNF signaling through TNFRSF1A prevented hepatic inflammation in Timp3−/− mice.
Mortality of Timp3−/− mice after hepatectomy
Given the liver pathology observed in unchallenged aged Timp3−/− mice, we reasoned that an acute challenge might further indicate the importance of TIMP3 in controlling TNF bioavailability. We examined the regenerative response after partial hepatectomy in mice aged 9–12 weeks, because partial hepatectomy induces a rapid release of TNF30. In addition, liver regeneration is dependent on signaling through TNFRSF1A and activation of NF-κB and IL-6 (ref. 27). Regeneration of the liver can be monitored by measuring liver mass, which is restored 8–10 d after partial hepatectomy in mice31.
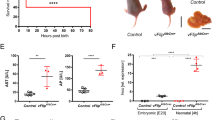
Liver to body mass ratios after partial hepatectomy increased similarly in wild-type and Timp3−/− mice until 72 h after partial hepatectomy (Fig. 5a). Beyond this time point, we observed a marked decline in liver mass of all Timp3−/− mice (Fig. 5a). By 144 h after partial hepatectomy, four of six Timp3−/− mice were moribund, and the remaining two, though not moribund, had decreased liver to body mass ratios. By contrast, all six wild-type mice appeared normal and had healthy liver to body mass ratios.
(a) Liver to body mass ratio over time after partial hepatectomy (PHx) in wild-type (dashed line) and Timp3−/− (solid line) mice. (b) Representative western blot of cyclin D1 and PCNA in regenerating livers of wild-type and Timp3−/− mice (50 μg of protein loaded per lane). (c–f) Immunohistochemical detection of phosphorylated histone H3 in regenerating livers of wild-type (c,e) and Timp3−/− (d,f) mice. Arrowheads indicate positive nuclei. (g) Histomorphometric quantification (original magnification, ×40) of nuclei positive for phosphorylated histone H3 in regenerating livers of wild-type and Timp3−/− mice. PHx, partial hepatectomy. Data are the mean ± s.e.m. (n = 3 and n = 6 mice of each genotype, aged 9–12 weeks, at time points 0–96 h and 144 h, respectively). *P < 0.05. Scale bars: c–f, 0.1 mm; insets in c–f, 20 μm.
Accelerated cell cycle in regenerating livers of Timp3−/− mice
In the resting liver, hepatocytes are in the G0 state31. After partial hepatectomy, a surge of TNF at 1 h triggers hepatocyte entry into the cell cycle30. We followed hepatocyte cell-cycle progression in Timp3−/− mice after partial hepatectomy by measuring cyclin D1 to mark the G1-S transition, proliferating cell nuclear antigen (PCNA) to indicate S phase, and phosphorylated histone H3 to quantify mitosis. Immunoblotting showed that cyclin D1 and PCNA were expressed earlier in the absence of TIMP3 than in wild-type mice (Fig. 5b). Hepatocyte cell division also occurred earlier in livers of Timp3−/− mice, as indicated by a substantial increase in the number of nuclei positive for phosphorylated histone H3 (21.1 ± 5.9% in Timp3−/− mice versus 1.8 ± 2.1% in wild-type mice) at 48 h (Fig. 5c–g). This level of mitosis was observed only at 72 h in wild-type mice (Fig. 5g). Taken together, these findings suggest that Timp3−/− hepatocytes rapidly complete the cell cycle.
Extensive necrosis of regenerating livers of Timp3−/− mice
We investigated the cause of mortality in Timp3−/− mice undergoing liver regeneration. During the window of hepatocyte cell-cycle progression (0–48 h), livers of Timp3−/− mice appeared normal and showed characteristics of regeneration, such as hepatocyte hypertrophy and perturbed architecture, similar to those of wild-type mice (Fig. 6a–c,g–i). By contrast, by 72 h Timp3−/− mice had developed multiple foci of liver necrosis (Fig. 6d,j), which became more marked with time (Fig. 6e,k). Histologically, necrosis was liquefactive in nature, and by 144 h no normal lobules could be seen, owing to widespread hepatocyte vacuolization (Fig. 6f,l).
Sustained activation of TNF leads to hepatocyte apoptosis
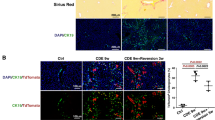
Because TNF can stimulate cell proliferation and induce programmed cell death13,32, we examined whether apoptosis accompanied hepatic necrosis in Timp3−/− mice. Histomorphometry after TUNEL staining detected numerous apoptotic nuclei by 48 h in regenerating livers of Timp3−/− but not wild-type mice (Fig. 7a–c). Molecular analyses indicated that there was a pronounced cleavage of BID at 24 h (Fig. 7d) and activation of caspase 3 at 48 h (Fig. 7e) after partial hepatectomy in Timp3−/− mice. α-tubulin was used as a loading control (Fig. 7f). These observations suggest that TNF-mediated hepatocyte apoptosis may initiate hepatocyte death, although other more complex mechanisms may contribute to the eventual collapse of liver regeneration in Timp3−/− mice.
(a) Quantification of TUNEL-positive nuclei in livers of wild-type and Timp3−/− mice after partial hepatectomy (PHx). (b,c) Representative TUNEL-stained liver sections from wild-type (b) and Timp3−/− (c) mice at 48 h after partial hepatectomy. Representative western blots of BID (d), caspase 3 (e) and α-tubulin (f) in regenerating livers of wild-type and Timp3−/− mice (50 μg of protein loaded per lane). Wild-type mouse liver treated with Fas agonist was used as a positive control for apoptosis markers (C), and α-tubulin was used as a protein loading control. (g,h) Quantification of TNF by ELISA in regenerating livers of Timp3−/− (g) and Timp1−/− (h) mice and their respective wild-type controls. (i) Quantification of IL-6 by ELISA in regenerating livers of wild-type and Timp3−/− mice. (j) Temporal expression of PPAR-γ (50 μg protein loaded per lane) in regenerating livers of wild-type and Timp3−/− mice. Data are the mean ± s.e.m. (n = 3 mice, aged 9–12 weeks, at each time point per group). PHx, partial hepatectomy. *P < 0.05. Scale bars, 40 μm.
One such mechanism may involve the upregulation of MMPs that are induced by TNF18. By real-time TaqMan RT-PCR, we found a substantial increase in the mRNA expression of Mmp9 at 72 h, Mmp13 at 1 h, Mmp14 between 24 and 96 h, and Mmp15 at 96 h after partial hepatectomy in livers of Timp3−/− mice relative to those of wild-type controls (data not shown). Gelatin zymography on liver extracts from regenerating livers of wild-type and Timp3−/− mice showed that there was an increase in MMP9 activity (data not shown). Notably, none of these MMPs had increased expression in livers of Timp3−/− mice before partial hepatectomy.
We investigated hepatic TNF levels over the time course of liver regeneration. As expected in wild-type mice, the level of TNF peaked sharply at 1 h after partial hepatectomy and then declined to baseline levels (Fig. 7g). Loss of TIMP3 function caused substantial perturbations in TNF throughout the course of liver regeneration: TNF was upregulated throughout the regeneration and did not decline as it did in wild-type mice (Fig. 7g). This effect was specific to Timp3−/− mice and was not observed in regenerating livers of Timp1−/− mice, which maintained a TNF expression profile similar to that of wild-type controls (Fig. 7h).
IL-6 is a key target of TNF signaling during liver regeneration27. We confirmed the perturbation of TNF signaling by assessing IL-6 levels by ELISA during the regenerative response. Consistent with higher levels of TNF, IL-6 levels were substantially higher and dysregulated in regenerating livers of Timp3−/− mice (Fig. 7i). Notably, both TNF and IL-6 were highest at 144 h after partial hepatectomy, coincident with the mortality of Timp3−/− mice (Fig. 7g,i). Peroxisome proliferator activated receptor-γ (PPAR-γ), a nuclear hormone receptor with an anti-inflammatory function33,34, is a marker of chronic cytokine exposure. We found a large increase in PPAR-γ at 48 h after partial hepatectomy in livers of Timp3−/− mice (Fig. 7j). These data suggest that livers of Timp3−/− mice have sustained TNF activity and inappropriate hepatocyte cell death.
TNF neutralizing antibody rescues Timp3−/− mice
We considered that TIMP3 is essential for controlling hepatic TNF activity and that excessive TNF is responsible for the untimely activation of hepatocyte cell death in regenerating livers of Timp3−/− mice. We therefore tested whether a neutralizing antibody to TNF could rescue these mice. Because TNF signaling in the first 6 h is required for hepatocyte DNA synthesis, we administered the neutralizing antibody or vehicle to Timp3−/− mice at 12 h after partial hepatectomy and examined livers at 96 h for organ pathology. Substantial necrosis was apparent in mice treated with vehicle alone (Fig. 8a–c). By contrast, all three mice treated with the TNF blocking antibody showed no signs of hepatic necrosis (Fig. 8d–f).
(a–f) Hematoxylin and eosin staining of regenerating livers of Timp3−/− mice treated with vehicle (phosphate-buffered saline) alone (a–c) or neutralizing antibody to TNF (d–f). A single dose was administered 12 h after partial hepatectomy and livers were collected at 96 h after partial hepatectomy. Arrowheads indicate necrotic areas. (g) Quantification of liver to body mass ratio at 96 h after partial hepatectomy in Timp3−/− mice treated with vehicle (gray bar) or neutralizing antibody (black bar). (h) Quantification of IL-6 by ELISA in regenerating livers of the Timp3−/− mice treated as above. Data are the mean ± s.e.m. (n = 3 mice, aged 9–12 weeks, per group). *P < 0.05, **P = 0.001. Scale bars, 0.1 mm.
Consistent with the rescued pathology, Timp3−/− mice had considerably higher liver to body mass ratios (Fig. 8g). We first determined hepatic TNF in these mice and found that TNF was reduced in mice treated with the neutralizing antibody (data not shown); however, we found that the antibody interfered with measurements of TNF by ELISA. Thus, to confirm the effective blocking of TNF activity, we measured IL-6 in liver homogenates by ELISA and found that hepatic IL-6 levels were 6.6 times lower in mice treated with the antibody (Fig. 8h). These results show that TNF dysregulation is responsible for the phenotype of Timp3−/− mice.
Discussion
We showed here that TIMP3 is a crucial innate negative regulator of TNF both in tissue homeostasis and in tissue response to injury. Timp3 deficiency leads to an increase in TACE activity and a failure to control the release of TNF, leading to inappropriate induction of inflammation and TNF-mediated cell death.
Previous studies have reported that, of the four members of the TIMP family, only TIMP3 inhibits TACE activity4. Shedding mediated by TACE extends the effects of TNF from juxtacrine to paracrine and provides an additional level of pericellular control of TNF activity. We found that deletion of Timp3 increased TACE activity in the liver, as shown by an increase in the cleavage of a TACE peptide substrate, which was inhibited only by recombinant TIMP3 and not by recombinant TIMP1. Notably, we also observed the constitutive appearance of the soluble 17-kDa TNF species, as well as an increase in levels of the 26-kDa membrane-bound form of TNF. Expression of Adam17 mRNA was not increased in livers of Timp3−/− mice before or during regeneration (data not shown), although there was a trend towards an increase in mature TACE protein in livers of Timp3−/− mice before partial hepatectomy (data not shown).
The activities of MMP7, the only other metalloproteinase shown to affect TNF release in vivo23, or of MMP14 and MMP17, which have been reported to cleave TNF in vitro21,22, were not elevated in livers of Timp3−/− mice before partial hepatectomy. These findings are consistent with the interpretation that TIMP3 is a physiological regulator of TACE activity and TNF shedding. TNF can induce its own expression in a positive-feedback manner, as well as the expression of several molecules including MMPs and TACE35. Whether changes in TNF or TACE expression in the absence of TIMP3 also contribute to an increase in TNF activity needs to be tested further.
Suppression of both basal and inducible TNF was compromised in Timp3−/− mice. Continuous, low-level exposure to TNF over the lifespan of Timp3−/− mice led to abnormal lymphocyte infiltration. This infiltration was prevented when Tnfrsf1a was deleted, indicating that this TNF signaling pathway is required for the observed pathology. Notably, lymphocytic infiltration is also observed in human livers afflicted with chronic active hepatitis28,29. We propose that TIMP3 constitutes a previously unknown point of control in TNF-mediated inflammation.
In addition to its ability to inhibit TACE, TIMP3 is also unique in that it binds to the extracellular matrix on secretion36. Notably, both TACE and TIMP3 are abundantly expressed in heart, kidney, lung, liver and brain17. In the liver, hepatocytes have been reported to be the primary source of TIMP3 (ref. 18). We confirmed this localization by in situ hybridization and found that livers of Timp3−/− mice containing wild-type bone marrow–derived cells expressed TIMP3 at low levels. Thus, TIMP3 is abundant in the liver, and its secretion by hepatocytes and Kupffer cells into the pericellular microenvironment positions it well for regulating TNF processing. The precise mechanism by which TIMP3 inhibits TACE and TNF release, however, requires further investigation. Similarly, the various biological effects of TNF shedding may be dependent on the context37,38,39 and need to be fully understood. Combining noncleavable transmembrane TNF transgenic expression with Timp3 deficiency will allow further investigation of the pathological consequences that arise from altered TNF shedding.
Macrophages are a principal source of TNF. In Timp3−/− mice, TNF was produced by Kupffer cells, the resident macrophage population of the liver. These cells are positioned to respond rapidly to infection or injury by initiating cytokine cascades. Maintenance of the normal noninflammatory state results not only from the absence of stimuli, but also from the active suppression of inappropriate inflammatory responses40. Tissue-specific inflammatory conditions spontaneously occur in several genetically altered mice40. The liver is one of the sites most frequently affected, probably owing to its large size and unique function40. Kupffer cells constitute the largest population of resident macrophages in the body41; are responsible for clearing gut-derived bacteria and lipopolysaccharide, a potent inducer of TNF42; and are a chief source of proinflammatory cytokines41. Kupffer cells therefore have an important role in liver tissue homeostasis and in the hepatic inflammatory response. It may be imperative to actively suppress an aberrant inflammatory reaction while Kupffer cells carry out their function. Thus, the presence of TIMP3 may be more crucial for liver tissue homeostasis in this context than in others.
The essential role of TIMP3 was more obvious after the induction of TNF signaling by partial hepatectomy, which resulted in massive hepatic necrosis, apoptosis and morbidity in Timp3−/− mice, owing to their inability to downregulate hepatic TNF levels. TNF is normally tightly regulated and is transiently induced after hepatectomy in wild-type mice. Levels of PPAR-γ were also increased in regenerating livers of Timp3−/− mice, which may be a reactive response to prolonged cytokine exposure and an attempt to negatively regulate cytokine production and macrophage activation33,34.
The dual role of TNF in cell survival and cell death is well recognized13,14 and was highlighted in our organ regeneration studies. At first, the increase in TNF accelerated hepatocyte cell-cycle progression in Timp3−/− mice, but then the sustained upregulation of TNF triggered programmed cell death and necrosis. Neutralizing antibody to TNF completely rescued hepatocyte cell death in Timp3−/− mice after partial hepatectomy. Consistent with the function of TIMP3 in controlling TNF, endogenous Timp3 was transcriptionally induced after the peak of TNF induction during liver regeneration in wild-type mice (data not shown). Our findings show that TIMP3 functions upstream of TNF. This metalloproteinase inhibitor alters both the amplitude and the duration of TNF activation, switching the effect of TNF from prosurvival to proapoptotic. Overall, TIMP3 regulation of TNF is essential for successful liver regeneration.
Inappropriate induction or sustained activation of TNF signaling has been implicated in the pathogenesis of numerous common human diseases14 and is recapitulated in TNF transgenic mouse models43,44. We provide evidence that TIMP3 is a crucial negative regulator of TNF release and signaling in vivo. We also show that TIMP3 has an essential role in hepatic inflammation and regeneration. The pathogenetic mechanisms leading to SFD, including the role of mutant TIMP3 proteins45,46, are currently not well understood. Macrophages and inflammation are reported to have a role in the development of age-related macular degeneration, which has a pathology closely resembling that of SFD47. It is plausible that TIMP3 is involved in the inflammation underlying age-related macular degeneration and possibly SFD. It will be important to explore the role of TIMP3 and inflammation in clinical conditions including age-related macular degeneration, cardiovascular disease, sepsis and rheumatoid arthritis.
Methods
Experimental mice.
We generated 129J/C57BL6 Timp3−/− mice as described2 and backcrossed them eight times into the C57BL/6 background. Unlike the original mixed strain, C57BL/6 Timp3−/− mice do not succumb to morbidity through lung dysfunction. We obtained C57BL/6 mice deficient in Tnfrsf1a (p55), originally generated by the laboratory of T.W. Mak (Ontario Cancer Institute), from Jackson Laboratory. We did PCR genotyping of Timp3 as described2. Primer sequences are available on request. We fed mice 4%-fat chow ad libitum and cared for them in accordance with protocols approved by the Ontario Cancer Institute Animal Care Committee and with Canadian Council on Animal Care guidelines.
Liver regeneration studies.
We used male Timp3−/− mice, 9–12 weeks old, and their age- and sex-matched wild-type littermates as controls. We anesthetized mice with isofluorane and carried out 70% partial hepatectomy as described48. We did the surgeries consecutively on knockout and control pairs. We killed mice by cervical dislocation and collected liver tissue. We flash-froze livers in liquid nitrogen for biochemical analyses, fixed them in 4% formaldehyde and embedded them in paraffin for histology.
ELISA assays.
We extracted total liver protein by homogenization of frozen tissue in extraction buffer containing 1% Triton X-100, 10 mM Tris-HCl (pH 7.6), 5 mM EDTA, 50 mM NaCl, 30 mM Na4P2O7, 50 mM NaF, 200 μM Na3VO4, 2 mM PMSF, 5 μg ml−1 of aprotinin, 1 μg ml−1 of pepstatin A and 2 μg of leupeptin. We centrifuged the suspension (at 14,000 r.p.m. for 20 min at 4 °C) and stored the supernatant at −70 °C. We determined protein concentration using a Dc BioRad assay. For each sample, we analyzed 4 μg of protein in triplicate by TNF and IL-6 ELISA kits used in accordance with the manufacturer's instructions (PharMingen).
Western-blot analyses.
We extracted liver proteins as described above for all western-blot analyses, except those for TNF and TACE, in which 1% SDS replaced 1% Triton X-100 in the buffer. We separated proteins by SDS-PAGE and transferred them to nitrocellulose membrane. We blocked membranes with 5% milk in TBST (20 mM Tris (pH 7.5), 500 mM NaCl and 0.05% Tween-20) and probed them with primary antibodies to TNF (Endogen), TACE (Chemicon), cyclin D1 (Oncogene), PCNA (Novacastra), BID (R&D Systems) or caspase 3 (Signal Transduction), followed by secondary antibodies to rabbit or to mouse (Cell Signaling) conjugated to horseradish peroxidase. We used enhanced chemiluminescence developing reagents from Pierce.
In situ hybridization.
We generated 35S-labeled riboprobes in both sense and antisense orientations from 300 bp of the Timp3 cDNA using T3 and T7 polymerases, respectively. We removed paraffin from serial 5-μm sections of liver and rehydrated them through a series of graded ethanol solutions, permeabilized them with proteinase K and postfixed them with 4% paraformaldehyde. We hybridized each section overnight at 50 °C with 2 × 106 c.p.m. of sense or antisense probe diluted in buffer for in situ hybridization (Dako). The next day, we washed the sections repeatedly at high stringency (0.1 × SSC, 55 °C), dehydrated them and dipped them in NBT-2 emulsion (Kodak). After 8 d of exposure, we developed the slides, counterstained them with Mayer's hematoxylin, mounted them and photographed them with bright-field and dark-field filters at × 40 magnification.
Bone marrow transplantation.
We isolated bone marrow from the tibia and femur of FVB mice universally expressing GFP driven by the chicken β-actin promoter (Jackson Laboratory). We removed red blood cells using Lympholyte-M (Cedarlane). We lethally irradiated 12-week-old FVB Timp3−/− mice with 900 rad and then injected them intravenously with 106 bone marrow–derived cells. To increase the number of bone marrow–derived Kupffer cells, we injected transplanted mice intraperitoneally with gadolinium chloride (20 mg per kg body weight in phosphate-buffered saline; Sigma). We killed mice 14 d after this treatment. Successful transplantation was confirmed by directly imaging freshly isolated spleen and liver by using fluorescence microscopy, followed by immunostaining with an antibody to GFP (Molecular Probes).
RNA preparation and real-time RT-PCR.
We prepared RNA from frozen tissues using Trizol reagent (Invitrogen) in accordance with the manufacturer's instructions. We designed oligonucleotide primers and fluorescently labeled TaqMan probes to recognize only full-length Timp3 RNA species by binding a region that is deleted in Timp3−/− RNA. We quantified gene expression as described17.
TACE activity assay.
We extracted liver proteins by homogenization of frozen liver tissue in buffer containing 50 mM Tris-HCl, 150 mM NaCl, 0.1% SDS, 1% sodium deoxycholate, 1% Nonidet P40 and 1% Triton X-100. We incubated 10 μM Abz-LAQAVRSSSR-Dpa (Peptides International) with 100 μg of liver lysates in TACE buffer (50 mM Tris-HCl (pH 7.4), 25 mM NaCl and 4% glycerol) at room temperature for 1 h. We measured fluorescence over 10 min with an excitation wavelength (λex) of 320 nm and an emission wavelength (λem) of 420 nm (ref. 49). We purchased recombinant human TIMP1 and human TIMP3 from Oncogene and R&D, respectively; BMS-275291 was a gift from R.A. Kramer (Bristol-Myers Squibb).
MMP activity assays.
We extracted liver proteins in the buffer described for ELISA excluding EDTA. We assayed 25-μl aliquots of liver homogenate at a protein concentration of 1 mg ml−1 in Tricine buffer (50mM Tricine (pH 7.4), 0.2 M NaCl, 10 mM CaCl2, 50 μM ZnSO4 and 0.005% Brij 35) containing 12 μl of 'Complete' EDTA-free protease inhibitor (Roche Diagnostics) and 0.8 mg of DQ-gelatin (Molecular Probes), 0.8 mg of DQ-fibronectin (Molecular Probes) or AEDENS/DABCYL-labeled KRPLGLARE peptide50. MMP7 activity was inhibited by the inclusion of 20 μM EDTA or 0.5 μM BMS-275291. We measured fluorescence at 37 °C as a function of time as follows: for DQ-gelatin and DQ-fibronectin, λex = 485 nm, λem = 538 nm; for KRPLGLARE, λex = 355 nm, λem = 538 nm. We purchased recombinant human MMP7 and human MMP17 from Calbiochem and human MMP14 from Chemicon.
Histological and immunohistochemical analyses.
We used 5-μm liver sections placed on Superfrost/Plus microscope slides (Fisher Scientific). We removed the paraffin from tissues in toluene, rehydrated them in water and ethanol and stained them with Harris' hematoxylin (Electron Microscopy Sciences) and eosin (Fisher Scientific) or with Masson's trichrome. We used antibodies to phosphorylated histone H3 (Upstate Biotech), TNF (PharMingen), IL-6 (PharMingen), p65 (Santa Cruz), F4/80 (CalTag), B220 (PharMingen), CD3 (Dako), CD4 (PharMingen), CD8 (PharMingen), mouse neutrophil (Serotec) and IFN-γ (Abcam) in accordance with the manufacturer's instructions. We used frozen sections for CD4 and CD8 immunostaining. We carried out TUNEL analysis with an ApoTag kit (Intergen). We quantified positive nuclei by blindly counting five random microscopic fields at × 40 magnification from two random sections for each experimental sample. Livers tested negative by Gram staining for Helicobacter hepaticus, which is associated with chronic hepatitis and hepatocellular neoplasia in mice.
TNF neutralizing experiment.
We administered 250 μl of monoclonal neutralizing antibody to TNF (1 mg ml−1; Genzyme) to mice intraperitoneally 12 h after partial hepatectomy. We killed mice 96 h after partial hepatectomy.
Statistical analyses.
Data are reported as the mean ± s.e.m. We carried out statistical analysis with Excel software (Microsoft). Comparisons were made by Student's t-test and analysis of variance.
References
Weber, B.H., Vogt, G., Pruett, R.C., Stohr, H. & Felbor, U. Mutations in the tissue inhibitor of metalloproteinases-3 (TIMP3) in patients with Sorsby's fundus dystrophy. Nat. Genet. 8, 352–356 (1994).
Leco, K.J. et al. Spontaneous air space enlargement in the lungs of mice lacking tissue inhibitor of metalloproteinases-3 (TIMP-3). J. Clin. Invest. 108, 817–829 (2001).
Fata, J.E. et al. Accelerated apoptosis in the Timp-3-deficient mammary gland. J. Clin. Invest. 108, 831–841 (2001).
Amour, A. et al. TNF-α converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 435, 39–44 (1998).
Lee, M.H., Knauper, V., Becherer, J.D. & Murphy, G. Full-length and N-TIMP-3 display equal inhibitory activities toward TNF-α convertase. Biochem. Biophys. Res. Commun. 280, 945–950 (2001).
Black, R.A. et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature 385, 729–733 (1997).
Moss, M.L. et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-α. Nature 385, 733–736 (1997).
Peschon, J.J. et al. An essential role for ectodomain shedding in mammalian development. Science 282, 1281–1284 (1998).
Killar, L., White, J., Black, R. & Peschon, J. Adamalysins. A family of metzincins including TNF-α converting enzyme (TACE). Ann. NY Acad. Sci. 878, 442–452 (1999).
Rovida, E., Paccagnini, A., Del Rosso, M., Peschon, J. & Dello Sbarba, P. TNF-α-converting enzyme cleaves the macrophage colony-stimulating factor receptor in macrophages undergoing activation. J. Immunol. 166, 1583–1589 (2001).
Contin, C. et al. Membrane-anchored CD40 is processed by the tumor necrosis factor-α-converting enzyme. Implications for CD40 signaling. J. Biol. Chem. 278, 32801–32809 (2003).
Lee, D.C. et al. TACE/ADAM17 processing of EGFR ligands indicates a role as a physiological convertase. Ann. N. Y. Acad. Sci. 995, 22–38 (2003).
Aggarwal, B.B. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 3, 745–756 (2003).
Chen, G. & Goeddel, D.V. TNF-R1 signaling: a beautiful pathway. Science 296, 1634–1635 (2002).
Gearing, A.J. et al. Processing of tumour necrosis factor-α precursor by metalloproteinases. Nature 370, 555–557 (1994).
McGeehan, G.M. et al. Regulation of tumour necrosis factor-α processing by a metalloproteinase inhibitor. Nature 370, 558–561 (1994).
Nuttall, R.K. et al. Expression analysis of the entire MMP and TIMP gene families during mouse tissue development. FEBS Lett. 563, 129–134 (2004).
Knittel, T. et al. Expression patterns of matrix metalloproteinases and their inhibitors in parenchymal and non-parenchymal cells of rat liver: regulation by TNF-α and TGF-β1. J. Hepatol. 30, 48–60 (1999).
Aldeguer, X. et al. Interleukin-6 from intrahepatic cells of bone marrow origin is required for normal murine liver regeneration. J. Hepatol. 35, 40–48 (2002).
Rizvi, N.A. et al. A phase I study of oral BMS-275291, a novel nonhydroxamate sheddase-sparing matrix metalloproteinase inhibitor, in patients with advanced or metastatic cancer. Clin. Cancer Res. 10, 1963–1970 (2004).
d'Ortho, M.P. et al. Membrane-type matrix metalloproteinases 1 and 2 exhibit broad-spectrum proteolytic capacities comparable to many matrix metalloproteinases. Eur. J. Biochem. 250, 751–757 (1997).
English, W.R. et al. Membrane type 4 matrix metalloproteinase (MMP17) has tumor necrosis factor-α convertase activity but does not activate pro-MMP2. J. Biol. Chem. 275, 4046–4055 (2000).
Haro, H. et al. LM. Matrix metalloproteinase-7-dependent release of tumor necrosis factor-α in a model of herniated disc resorption. J. Clin. Invest. 105, 143–150 (2000).
Ghosh, S., May, M.J. & Kopp, E.B. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16, 225–260 (1998).
Beg, A.A., Sha, W.C., Bronson, R.T., Ghosh, S. & Baltimore, D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature 376, 167–170 (1995).
Alcamo, E. et al. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-κB in leukocyte recruitment. J. Immunol. 167, 1592–1600 (2001).
Yamada, Y., Kirillova, I., Peschon, J.J. & Fausto, N. Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc. Natl. Acad. Sci. USA 94, 1441–1446 (1997).
Jevon, G.P. Grade and stage of chronic hepatitis. Pediat. Dev. Pathol. 4, 372–380 (2001).
Toyonaga, T. et al. Chronic active hepatitis in transgenic mice expressing interferon-γ in the liver. Proc. Natl. Acad. Sci. USA 91, 614–618 (1994).
Michalopoulos, G.K. & DeFrances, M.C. Liver regeneration. Science 276, 60–66 (1997).
Fausto, N. Liver regeneration. J. Hepatol. 32, 19–31 (2000).
Kyriakis, J.M. Life-or-death decisions. Nature 414, 265–266 (2001).
Ricote, M., Li, A.C., Willson, T.M., Kelly, C.J. & Glass, C.K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature 391, 79–82 (1998).
Jiang, C., Ting, A.T. & Seed, B. PPAR-γ agonists inhibit production of monocyte inflammatory cytokines. Nature 391, 82–86 (1998).
Bzowska, M., Jura, N., Lassak, A., Black, R.A. & Bereta, J. Tumor necrosis factor-α stimulates expression of TNF-α converting enzyme in endothelial cells. Eur. J. Biochem. 271, 2808–2820 (2004).
Blenis, J. & Hawkes, S.P. Characterization of a transformation-sensitive protein in the extracellular matrix of chicken embryo fibroblasts. J. Biol. Chem. 259, 11563–11570 (1984).
Georgopoulos, S., Plows, D. & Kollias, G. Transmembrane TNF is sufficient to induce localized tissue toxicity and chronic inflammatory arthritis in transgenic mice. J. Inflamm. 46, 86–97 (1996).
Mueller, C. et al. Noncleavable transmembrane mouse tumor necrosis factor-α (TNFα) mediates effects distinct from those of wild-type TNFα in vitro and in vivo. J. Biol. Chem. 274, 38112–38118 (1999).
Ruuls, S.R. et al. Membrane-bound TNF supports secondary lymphoid organ structure but is subservient to secreted TNF in driving autoimmune inflammation. Immunity 15, 533–543 (2001).
Nathan, C. Points of control in inflammation. Nature 420, 846–852 (2002).
Bouwens, L. Structural and functional aspects of Kupffer cells. Revis. Biol. Celular 16, 69–94 (1988).
Dory, D. Generation and functional characterization of a clonal murine periportal Kupffer cell line from H-2Kb–tsA58 mice. J. Leukoc. Biol. 74, 49–59 (2003).
Kontoyiannis, D., Pasparakis, M., Pizarro, T.T., Cominelli, F. & Kollias, G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10, 387–398 (1999).
Butler, D.M. et al. DBA/1 mice expressing the human TNF-α transgene develop a severe, erosive arthritis: characterization of the cytokine cascade and cellular composition. J. Immunol. 159, 2867–2876 (1997).
Yeow, K.M. et al. Sorsby's fundus dystrophy tissue inhibitor of metalloproteinases-3 (TIMP-3) mutants have unimpaired matrix metalloproteinase inhibitory activities, but affect cell adhesion to the extracellular matrix. Matrix Biol. 21, 75–88 (2002).
Qi, J.H. et al. Expression of Sorsby's fundus dystrophy mutations in human retinal pigment epithelial cells reduces matrix metalloproteinase inhibition and may promote angiogenesis. J. Biol. Chem. 277, 13394–13400 (2002).
Ambati, J. et al. An animal model of age-related macular degeneration in senescent Ccl-2- or Ccr-2-deficient mice. Nat. Med. 9, 1390–1397 (2003).
Higgins, G.M. & Anderson, R.M. Restoration of the liver of the white rat following partial surgical removal. Arch. Pathol. 12, 186–182 (1931).
Jin, G. et al. A continuous fluorimetric assay for tumor necrosis factor-α converting enzyme. Anal. Biochem. 302, 269–275 (2002).
McIntyre, J.O. et al. Development of a novel fluorogenic proteolytic beacon for in vivo detection and imaging of tumor-associated matrix metalloproteinase-7 activity. Biochem. J. 377, 617–628 (2004).
Acknowledgements
We thank L. Salmena and R. Hakem for the antibodies for apoptosis analyses, G.A. Wood for critically reading the manuscript, B. Anand-Apte for discussion, and P. Waterhouse for advice on experimental design. This work was supported by grants from the Canadian Institute of Health Research to R.K.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Mohammed, F., Smookler, D., Taylor, S. et al. Abnormal TNF activity in Timp3−/− mice leads to chronic hepatic inflammation and failure of liver regeneration. Nat Genet 36, 969–977 (2004). https://doi.org/10.1038/ng1413
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1413
This article is cited by
-
Transcriptomic landscape of TIMP3 oncosuppressor activity in thyroid carcinoma
Cancer Cell International (2022)
-
Treatment of Sorsby fundus dystrophy with anti-tumor necrosis factor-alpha medication
Eye (2022)
-
Tumor necrosis factor-α and matrix metalloproteinase-9 cooperatively exacerbate neurovascular degeneration in the neonatal rat retina
Cell and Tissue Research (2022)
-
Poncirin attenuates CCL4-induced liver injury through inhibition of oxidative stress and inflammatory cytokines in mice
BMC Complementary Medicine and Therapies (2020)
-
Post-stroke inflammation—target or tool for therapy?
Acta Neuropathologica (2019)